HicAggR - Quick start
Nicolas Chanard, David Depierre & Olivier Cuvier
2024-07-07
Source:vignettes/HicAggR.Rmd
HicAggR.RmdQuick start from the import of HiC data to the aggregation of HiC contacts. It includes 4 steps:
- Import HiC
- Import genomic coordinates
- Submatrices extractions
- Plot and visualization
Test dataset
Description
Data were obtained from Drosophila melanogaster S2 cells. HiC test dataset: Directly downloaded from the 4DN platform. Genomic coordinates: ChIPseq peaks of Beaf-32 protein in wild type cells (GSM1278639).
Genomic 3D structure
For a test, please download HiC data in .hic format (Juicer).
withr::local_options(list(timeout = 3600))
cache.dir <- paste0(tools::R_user_dir("", which="cache"),".HicAggR_HIC_DATA")
bfc <- BiocFileCache::BiocFileCache(cache.dir, ask = FALSE)
if(length(BiocFileCache::bfcinfo(bfc)$rname)==0 ||
!"Control_HIC.hic"%in%BiocFileCache::bfcinfo(bfc)$rname){
Hic.url <- paste0("https://4dn-open-data-public.s3.amazonaws.com/",
"fourfront-webprod/wfoutput/7386f953-8da9-47b0-acb2-931cba810544/",
"4DNFIOTPSS3L.hic")
if(.Platform$OS.type == "windows"){
HicOutput.pth <- BiocFileCache::bfcadd(
x = bfc,rname = "Control_HIC.hic",
fpath = Hic.url,
download = TRUE,
config = list(method="auto",mode="wb"))
}else{
HicOutput.pth <- BiocFileCache::bfcadd(
x = bfc,rname = "Control_HIC.hic",
fpath = Hic.url,
download = TRUE,
config = list(method="auto"))
}
}else{
HicOutput.pth <- as.character(BiocFileCache::bfcpath(bfc)[
which(BiocFileCache::bfcinfo(bfc)$rname=="Control_HIC.hic")])
}Genomic location and annotation data
These kind of data can be imported in R with rtracklayer package.
data("Beaf32_Peaks.gnr")View
| seq | start | end | strand | name | score |
|---|---|---|---|---|---|
| 2L | 35594 | 35725 | * | Beaf32_2 | 76 |
| 2L | 47296 | 47470 | * | Beaf32_3 | 44 |
| 2L | 65770 | 65971 | * | Beaf32_5 | 520 |
Additional genome informations
Required genomic information used by the functions during the entire
pipeline are a data.frame containing chromosomes names and
sized and the binSize, corresponding to the HiC matrices at
the same resolution.
seqlengths.num <- c('2L'=23513712, '2R'=25286936)
chromSizes <- data.frame(
seqnames = names(seqlengths.num ),
seqlengths = seqlengths.num
)
binSize <- 10001 Import HiC
The package supports the import and normalization of HiC data.
NOTE: Since version 0.99.2, the package supports import of balanced HiC matrices in .hic, .cool/.mcool formats. It also supports the import of ‘o/e’ matrices in .hic format.
Import
HicAggR can import HiC data stored in the main formats: .hic, .cool, .mcool, .h5. The pacakage imports by default the raw counts in R. Therefore, it is necessary to perform the balancing and observed/expected correction steps.
Balancing
hicLst <- BalanceHiC(hicLst)2 Import genomic coordinates
Genomic coordinates data (ChIP seq peaks or any other feature) need to be indexed using the same referenced genome as for HiC data. Then the genomic coordinates are paired in GInteraction objects.
Features Indexing
Beaf_Index.gnr <- IndexFeatures(
gRangeList = list(Beaf = Beaf32_Peaks.gnr),
chromSizes = chromSizes,
binSize = binSize
)Beaf32 <-> Beaf32 putatives
Constraints for the distance between interaction sites are defined here in order to limit the number of pairs.
Beaf_Pairs.gni <- SearchPairs(
indexAnchor = Beaf_Index.gnr,
minDist = "25KB",
maxDist = "100KB"
)3 Submatrices extractions
Once data have been imported, interactions are extracted out of the pairs of genomic coordinates.
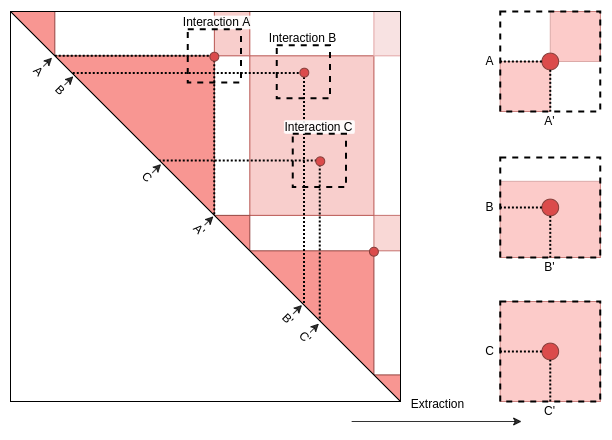
Extraction
Beaf.mtx_lst <- ExtractSubmatrix(
genomicFeature = Beaf_Pairs.gni,
hicLst = hicLst
)4 Plot and visualization
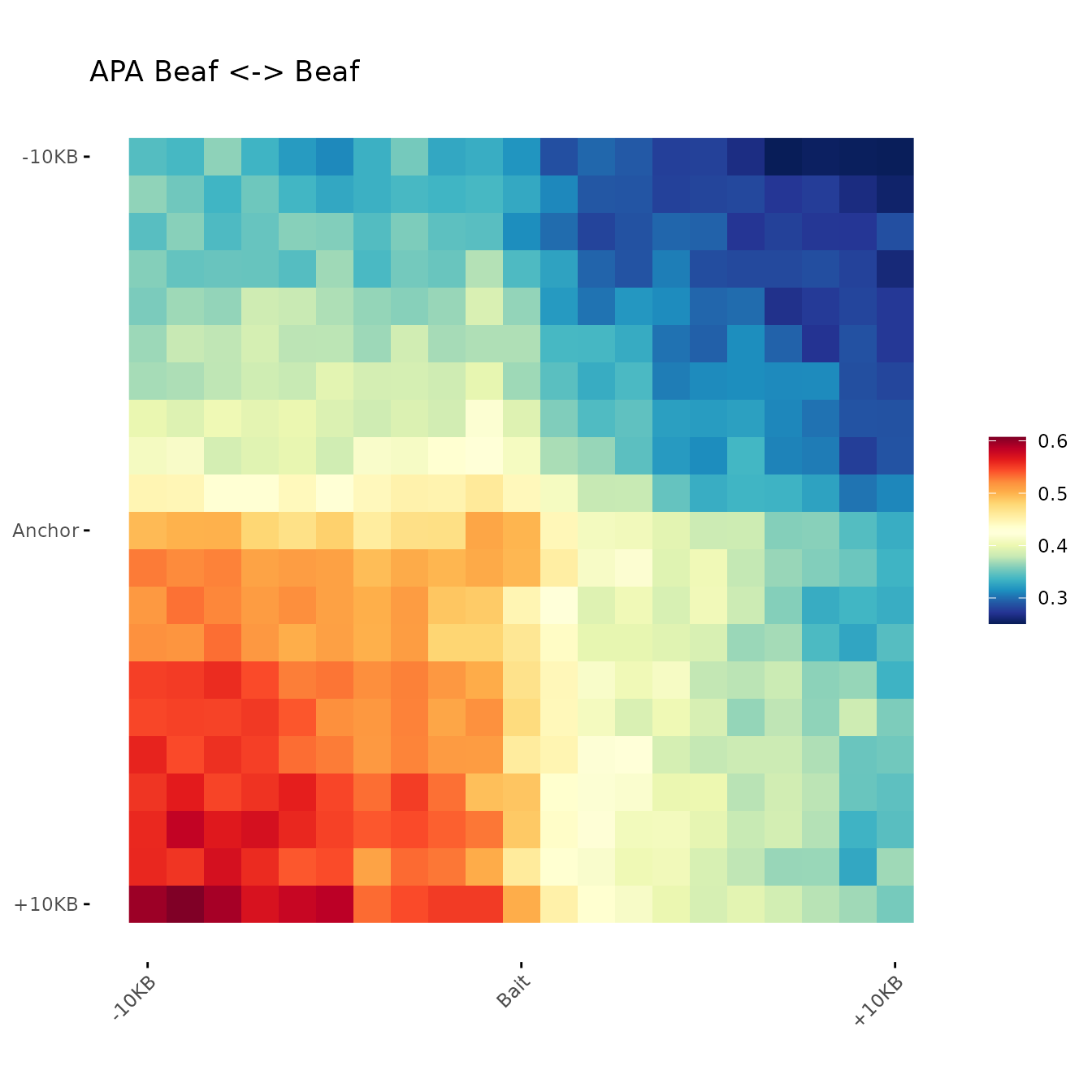
Submatrices are aggregated as sum, average or median. Then, aggregated matrix is plotted as a heatmap of contact frequencies (in the example, contacts surounding Beaf-32 sites).
Prepare matrices list
Beaf.mtx_lst <- PrepareMtxList(
matrices = Beaf.mtx_lst
)Aggregation
aggreg.mtx <- Aggregation(Beaf.mtx_lst)Session Info
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.4 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] HicAggR_1.1.3
#>
#> loaded via a namespace (and not attached):
#> [1] tidyselect_1.2.1 farver_2.1.2
#> [3] dplyr_1.1.4 blob_1.2.4
#> [5] filelock_1.0.3 fastmap_1.2.0
#> [7] reshape_0.8.9 BiocFileCache_2.12.0
#> [9] digest_0.6.36 lifecycle_1.0.4
#> [11] RSQLite_2.3.7 magrittr_2.0.3
#> [13] compiler_4.4.1 rlang_1.1.4
#> [15] sass_0.4.9 tools_4.4.1
#> [17] utf8_1.2.4 yaml_2.3.9
#> [19] data.table_1.15.4 knitr_1.47
#> [21] labeling_0.4.3 S4Arrays_1.4.1
#> [23] bit_4.0.5 curl_5.2.1
#> [25] DelayedArray_0.30.1 plyr_1.8.9
#> [27] abind_1.4-5 BiocParallel_1.38.0
#> [29] withr_3.0.0 purrr_1.0.2
#> [31] BiocGenerics_0.50.0 desc_1.4.3
#> [33] grid_4.4.1 stats4_4.4.1
#> [35] fansi_1.0.6 colorspace_2.1-0
#> [37] Rhdf5lib_1.26.0 ggplot2_3.5.1
#> [39] scales_1.3.0 SummarizedExperiment_1.34.0
#> [41] cli_3.6.3 rmarkdown_2.27
#> [43] crayon_1.5.3 ragg_1.3.2
#> [45] generics_0.1.3 httr_1.4.7
#> [47] DBI_1.2.3 cachem_1.1.0
#> [49] rhdf5_2.48.0 stringr_1.5.1
#> [51] zlibbioc_1.50.0 parallel_4.4.1
#> [53] XVector_0.44.0 matrixStats_1.3.0
#> [55] vctrs_0.6.5 Matrix_1.7-0
#> [57] jsonlite_1.8.8 IRanges_2.38.1
#> [59] S4Vectors_0.42.1 bit64_4.0.5
#> [61] systemfonts_1.1.0 tidyr_1.3.1
#> [63] strawr_0.0.91 jquerylib_0.1.4
#> [65] glue_1.7.0 pkgdown_2.1.0
#> [67] codetools_0.2-20 stringi_1.8.4
#> [69] gtable_0.3.5 GenomeInfoDb_1.40.1
#> [71] GenomicRanges_1.56.1 UCSC.utils_1.0.0
#> [73] munsell_0.5.1 tibble_3.2.1
#> [75] pillar_1.9.0 htmltools_0.5.8.1
#> [77] rhdf5filters_1.16.0 GenomeInfoDbData_1.2.12
#> [79] R6_2.5.1 dbplyr_2.5.0
#> [81] textshaping_0.4.0 evaluate_0.24.0
#> [83] lattice_0.22-6 Biobase_2.64.0
#> [85] highr_0.11 backports_1.5.0
#> [87] memoise_2.0.1 bslib_0.7.0
#> [89] Rcpp_1.0.12 InteractionSet_1.32.0
#> [91] gridExtra_2.3 SparseArray_1.4.8
#> [93] checkmate_2.3.1 xfun_0.45
#> [95] fs_1.6.4 MatrixGenerics_1.16.0
#> [97] pkgconfig_2.0.3